Professor Daisuke Kihara models novel coronavirus proteins
10-21-2020
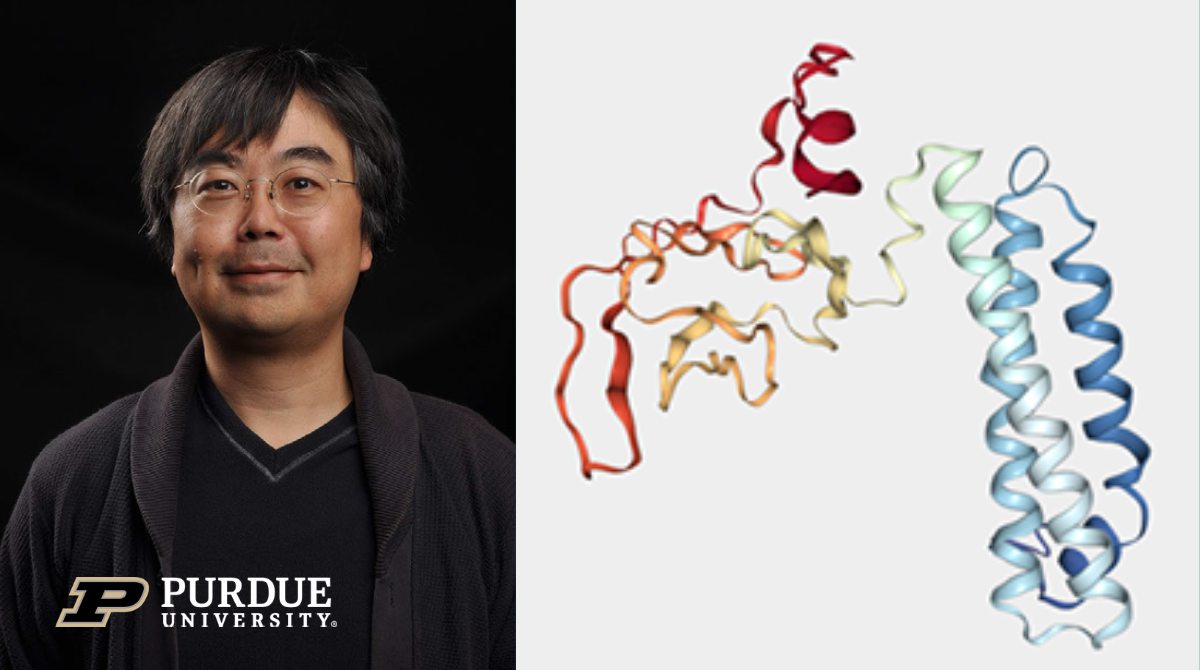
Professor Daisuke Kihara (Department of Computer Science and Biological Science) and researchers in Kihara Lab have used Purdue’s supercomputers to develop computational models of the proteins in the novel coronavirus SARS-CoV-2 that may prove useful for the development of drugs to treat COVID-19.
Experimental techniques like cryo-electron microscopy can provide a more definitive picture of a protein’s structure, but computational methods are much faster – something that is important for a fast-spreading virus like this where time is of the essence.
“Without anything in front of you, it’s difficult to think about how to develop a drug,” says Kihara. “But if you have a model of the structure, then information can come together that will let you generate a hypothesis and go from there on drug development.”
Kihara uses Gilbreth, Purdue’s powerful GPU cluster optimized for machine learning applications, to compare the protein’s amino acid sequence to sequences in a database of known proteins from other viruses and predict which pairs of amino acids are interacting or not. He then uses the Brown and Halstead clusters to run the simulation structure model and build a three-dimensional model of the protein.
In addition to his work on the novel coronavirus proteins, Kihara has also used Purdue’s supercomputers for his success in the CASP (Critical Assessment of protein Structure Prediction) and CAPRI (Critical Assessment of PRediction of Interactions) protein modeling contests. In 2014 and 2016, Kihara and his team used donated nodes from the clusters and were among a handful of top finishers chosen to present at CASP meetings to assess the results. They are also consistently ranked among the top in the CAPRI contests. They used nodes from the Halstead cluster for the 2020 contests.
Writer: Adrienne Miller, science and technology writer, Information Technology at Purdue (ITaP), 765-496-8204, mill2027@purdue.edu.